In August we started the project only focus on Spain and Madagascar. After extracting the GBIF data that was less than 3 Million records and around 800 protected areas (polygons). With this "little" data I thought I could use some brute force and did not spend much time optimizing the processing of the data on PostGIS. In total it needed around 8 hours to analyze all data and place it in a DB schema that was enough for the service layer of the widget. These scripts are available here.
The project was very well received and soon we were asked to extend the analysis to all countries, all protected areas, and all GBIF data. That is around 90.000 protected areas and 130 million occurrences from GBIF. We knew from the beginning that the previous strategy would not scale to handle so much data and that the strategies for data representation would also not work with protected areas of the size of Spain and millions of occurrences inside. So this time we decided to go in two different ways at the same time: Using Hadoop to do the spatial join and adapt the PostGIS strategy to make it more scalable. This post is of course talking about the second strategy.
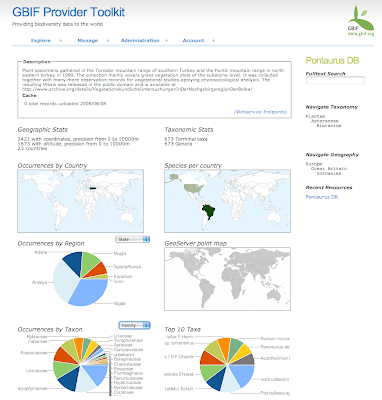
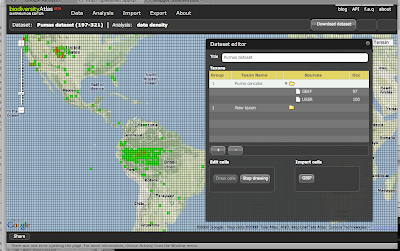
We wanted to work a little bit generic here and be able to support any kind of "polygon" analysis. That mean we would not be protected areas specific. So we have defined a Starting schema and a target schema databases. The starting schema is how we expect to get the data at the beginning and the target schema is the final processed schema that will be used to serve the data to the widget. GBIF gave us the primary data and WCMC the World Database on Protected Areas. The starting schema looks like:

The data from GBIF was given as a MySQL dump and imported into PostgreSQL using COPY statements. The WDPA on the other hand was given as a huge shapefile that we import using shp2pgsql.
The main things we needed to process were stats on:
The numbers in red are the number of records on each table after the data was processed.- Find which GBIF occurrences are inside which polygons
- Which providers have data for each polygons
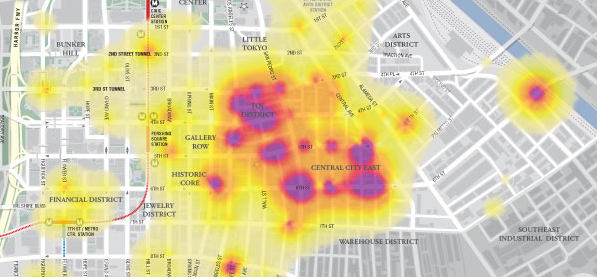
- Gridify the occurrences to be able to visualize them dynamically on Google Maps.
- Construct a taxonomic tree for each polygon and grid.
So the target schema looks like this (click to expand):
This is the definition of main tables:
occurrence: A primary data record. The location of a species at a certain place in a certain moment.
site: Even if there are 130M occurrences they are only in around 3.5M distinct points. A site is a point on earth where an occurrence is. This just allows us to geoprocess only the distinct positions and save us lot of time.
grid10,grid5...: The grouping of the sites in cells at different scales, 10, 5,1,0.5 and 0.1 degree. After 0.1 we go dynamically grouping using PostGIS SnaptoGrid (another full post about it would be needed).
The whole processing is done using PostgreSQL and PostGIS with some new functions that we needed to create. The details about the whole process is described here . And the main steps are:
1) Adjust PostgreSQL: use 2GB of RAM. AutoVacuum Off. Use as many processors as possible.
2) Get the sites: No more than a grouping of the occurrences by coordinates.
3) Spatially join the sites with the polygon table: This is where it gets interesting. The fastest way we found was using the distance function and run it threaded in parallel in different parts of the site. This is a computer intensive task so I used the 8 cores in my computer. The task takes around 4 hours. But depend s a lot on the complexity of the polygons and how good you are at distributing the load among different processors. We used a PHP script and a shell script to run several queries at the same time.
4) Delete sites that are not inside polygons and later delete the occurrences that are not in sites.
5) Create the grids: we are using our own grid system based on work previously done by Tim Robertson. The way we group the sites on the occurrences is by spatially joining the sites against tables with all the grid cells pre-created. To generate the grids Tim developed a little Java tool that create PostGIS insert statements. It is the GridBuilder class . But if someone needs them we can provide them directly as zip SQL files.
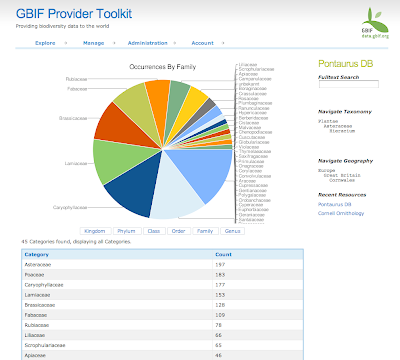
6) Generate stats: Calculate the number of occurrences, species, specimens, observations, other basis of records, plants, animals and other kingdoms. Those stats are available per polygon, per grid, per site, per provider, per taxon, etc... Lot of queries but that run very fast.
7) Generate taxonomy for each polygon: this can be a complicate process. The idea is to generate classifications per polygon based on the occurrences that are inside these polygons. We have created a PostgreSQL function that scan the occurrence table per polygon and generate the classification. The source code of this function is here .
And this is more or less everything. Of course lot of details are skipped here, like when to vacuum, generate indexes, etc. But you can find most of these details on the description page of this strategy .
You can find in any case all the source code for all those things on the Google Code GBIF-WDPA project. There you will find the source code of the Java SOA application, the Flex source code of the widget, the processing scripts and comments, well, everything created during this project.
we are working on a single script that will run the full process in one shot and it will be available there when ready.
Summary:
Well, after this long post my main recomendations if you need to use PostGIS to do something similar would be:
-Adjust postgreSQL settings regarding memory.
-Consider threading long operations that require a huge table scan.
-"Create table as" is much faster than updating.
-Amazon Web Services can be very slow due to I/O performance issues.
-Buy a Wii to play while you wait for results :)
Leave us a comment if you have any question or suggestions regarding this processing strategy.
And finally happy christmas everybody!