Tuesday, December 15, 2009
Recreating the European Starling story
Specially appealing is the story on how it got introduced in North America. I will just quote Wikipedia for that:
Although there are approximately 200 million starlings in North America, they are all descendants of approximately 60 birds (or 100 [1]) released in 1890 in Central Park, New York, by Eugene Schieffelin, who was a member of the Acclimation Society of North America reputedly trying to introduce to North America every bird species mentioned in the works of William Shakespeare.
I knew this was a story that could really be catchy. Specially if we could use scientific primary data to show this story. While working with Tim Robertson and Andrew Hill we started thinking about using Clustr, from Flickr, to create polygons out of primary data and see if we could display this story. I demoed this in Geoweb and TDWG this year and the feedback was most of the time really good. You can watch the video at Vimeo.
The challenge for that was that there is more than 1 million observations of the starling now available on GBIF and the classical point in map did not work well, the visualizations were tedious... well, kind of complicate. But the second semester of this year we started to see interactive maps that seemed to be analyzing raster images on the fly in Flash. This is really really cool. And since then we were just thinking more and more in raster representation of data to further filter in the client and allowing much more rich story telling. And then, one day, I showed the work from Andrew Cottam from WCMC on sea level rise and Google Maps for Flash. That was awesome! And being such a nice guy he is, he publish his code and saved me the time of figuring out the bitwise operations needed for at least one band raster. I am not sure if he wants me to put a link to his ongoing work so I will wait for him to publish it first (maybe in this blog ;) ).
So I could not resist and with the help of Tim preparing the raster tiles for the starling, and Sergio doing some UI, we prepared the following demo application.
(Click the image to open)
Drag the slider from 1880 to 2010 to see the accumulative records (by date recorded) for the data available on the GBIF network. While you drag the slider you will be presented with tooltips mostly taken from Wikipedia.
Soon we will release all the source code, once a bit cleaned, and will share more technical details. And the best is yet to come... we only used one band on this demo, but we have 3 to play with!!
I hope you like it and want to share some comments.
Ah! Dont forget to turn on sound!
Monday, November 16, 2009
Amazon EC2, EBS RAID-0 & PostGIS build script
Monday, November 9, 2009
Automated informatics pipelines, public datasets, and the cloud
A couple of days ago I became curious if I could use CloudBurst to compare novel influenza sequences to the entire known sequence record of influenza in the hopes of detecting novel reassortants on the fly. The intended use of CloudBurst is to take small sequence reads from high throughput sequencing projects and query them against a complete genome a variety of reasons including SNP discovery, genotyping and personal genomics1. So why not take a novel influenza and break it up into many small overlapping fragments and query them against a genome comprised of all known sequences to detect reassortment?
Comparisons similar to these can likely be done using some manipulation of the Blast or similar algorithm. But in my case, I was particularly interested in CloudBurst because of its available implementation in EC2 using Hadoop to utilise the cluster environment. This means that if successful I could later wrap my method into an automated workflow for detecting and reporting potentially novel influenza reassortants as soon as new sequences are reported.
I posted an overview of the steps it took below. In the end, as a first pass detection of reassortment it appears promising or at least interesting. All in all, I went from idea, to data assembly, to trial implementation in about 2 hours (not counting a few hours trying to get Hadoop to run the CloudBurst.jar. Hadoop version number = VERY important). That is great for the study of influenza, but what about biodiversity informatics?
There are a few things that made this so simple.
First, a rapid adoption of EC2 by the bioinformatics community. Look for example at the JVCI maintained Bio-linux distribution for use on EC2. It comes prepackaged with a whole smörgåsbord of very fun little tools. It costs 34 cents per hour (not 10 because it requires a 64-bit instance) and takes about 1 minute to set up your very own copy. Where is our (the biodiversity informatics community's) GISing, niche modeling, distribution mapping, PDing, SGAing, Hadoop spatial joining, machine image?
Second, a growing community of users who are exchanging knowledge about how to tackle these very large datasets in the cloud. Biodiversity informatics is growing its own such community, writers of this blog can attest to that.
Third, cloud hosted datasets. Amazon Web Services is making it fairly simple to host very large datasets that anyone in the community can access. The GenBank image is ~200gb and takes 5 minutes to set up as a mounted volume for my own use. This targets a completely different consumer then our data portals and APIs. It opens up the data for the community of informatics enthusiasts doing cool things on the cloud (think spelling correction, retrospective georeferencing, spatial joining).
With all that said I'd like to return to think about how this relates to biodiversity informatics. I've been talking with a variety of people over the past few weeks and all of them show some level of interest in moving to the cloud. What I'm worried about now is that we will adopt many of the same stances as a community as we have had without the cloud. The case study I developed above relied heavily on easily accessible data and tools. We as a community must move forward with this as a primary goal. Likely, there will always be a need for portals and APIs, but for really big questions sometimes it is just easier to have the entire dataset ready for access. Why hasn't our community got it together enough to launch a unified public dataset in the cloud?
I guess data quality concerns and ownership are two primary concerns. I'm sorry, but those are bad reasons, time to grow up, it isn't 2001 any longer.
Once in place we can begin to build new methods (or reapply existing ones) for parsing out duplicate records, linking data to geographic areas, merging error types into targeted datasets, and sharing findings with the owners of the original data. The 'snapshot' approach as implemented in AWS Public Data Set makes it so we consumers constantly rely on the original providers to include the newest and most up to date records, none of our hard work will be included in future snapshots if we don't come up with methods for reporting corrections to the source.
It is important to restate, I don't think portals and APIs will go away. They are for two completely different consumer communities than those interested in looking at the entire dataset at once, versus pulling smaller subsets of data manually or developing tools to do so via API interfaces. I do think that by providing public datasets, new methods and technologies for enhancing the portals and APIs will arise, as well as still unknown methods for improving the datasets at source, and ultimately enhancing our knowledge about the world's biodiversity.
After downloading Hadoop (version 0.18.3), here is what I ran.
Launch Hadoop cluster, create and mount a volume containing the GenBank influenza data
>src/contrib/ec2/bin/hadoop-ec2 launch-cluster cloud 5
>ec2-create-volume --snapshot snap-fe3ec297 -z us-east-1d
>ec2-attach-volume vol-7b49a612 -i
MASTERNODEIDHERE -d /dev/sdh >src/contrib/ec2/bin/hadoop-ec2 login cloud
$mkdir /inf
$mount /dev/sdh /inf
Here I ran a small Python script to extract all sequences (hemagluttanin only) known prior to the original swine flu outbreak, concatenate those sequences into a single “genome” saved as data/genome.fa, and recording a map of where in the genome each sequence ended.
Next, a group of sequences (again, hemagluttanin only) from the earliest swine flu outbreaks were broken up into numerous overlapping fragments and saved as data/segs.fa, again keeping a map of where each fragment belonged. These are the 'novel sequences' I would test for any cases of reassortment.
Download and extract CloudBurst
$wget http://downloads.sourceforge.net/project/cloudburst-bio/cloudburst/CloudBurst-1.0.1/CloudBurst-1.0.1.tgz
$tar xzf CloudBurst-1.0.1.tgz
$mv CloudBurst-1.0.1/ data/
Convert fasta files to Hadoop ready files
$java -jar data/ConvertFastaForCloud.jar data/genome.fa data/genome.br
$java -jar data/ConvertFastaForCloud.jar data/segs.fa data/segs.br
Move the data to hdfs
$/usr/local/hadoop-0.17.0/bin/hadoop fs -put ~/data /data
Run the analysis
$/usr/local/hadoop-0.17.0/bin/hadoop jar ~/data/CloudBurst.jar /data/genomeA.br /data/segsA.br /results 40 3 0 1 50 15 5 5 128 5
Copy results from hdfs
$/usr/local/hadoop-0.17.0/bin/hadoop fs -get /results results
Convert the results back to human readable format
java -jar data/PrintAlignments.jar results >results.txt
Profit!
Parsing meaning out of the results was a bit more labor intensive and I forewent any automation (for now), using OpenOffice spreadsheets to map matched portions back to their original viruses and report accession numbers. What I found was interesting, exciting and promissing, even matching some of the findings reported in Kingsford et al., 2009, from only 2 hours of work and a couple of dollars. I was pretty satisfied.
Wednesday, October 21, 2009
Install PostgreSQL 8.4 and PostGIS 1.4.0 in Ubuntu 9.0.4
Just for the record. The EC2 instance I used was ami-ccf615 from http://alestic.com .
Once login (totally fresh).
apt-get update apt-get install vim
#The sources are still not available on the regular package servers... edit the sources vim /etc/apt/sources.list add deb http://ppa.launchpad.net/pitti/postgresql/ubuntu jaunty main deb-src http://ppa.launchpad.net/pitti/postgresql/ubuntu jaunty main sudo apt-key adv --keyserver keyserver.ubuntu.com --recv-keys 8683D8A2 sudo apt-get update sudo apt-get install postgresql-8.4
#This changes the port from 5433 to 5432 sudo sed -i.bak -e 's/port = 5433/port = 5432/' /etc/postgresql/8.4/main/postgresql.conf sudo /etc/init.d/postgresql-8.4 stop sudo /etc/init.d/postgresql-8.4 start apt-get install postgresql-server-dev-8.4 libpq-dev apt-get install libgeos-dev wget http://postgis.refractions.net/download/postgis-1.4.0.tar.gz apt-get install proj tar xvfz postgis-1.4.0.tar.gz cd postgis-1.4.0 ./configure make make install sudo su postgres
#change the postgres password to "atlas" so that you can later login psql -c"ALTER user postgres WITH PASSWORD 'atlas'" createdb geodb (with password atlas) createlang -dgeodb plpgsql psql -dgeodb -f /usr/share/postgresql/8.4/contrib/postgis.sql psql -dgeodb -f /usr/share/postgresql/8.4/contrib/spatial_ref_sys.sql psql -dgeodb -c"select postgis_lib_version();"
#This should return 1.4.0exitSunday, September 20, 2009
Using Geowebcache tiles stored in s3 from Google Maps for Flash
Friday, August 14, 2009
Presentation at Geoweb 09: Biodiversity: Where is important

Friday, July 3, 2009
Biodiversity databases and OGC standards don't play well together

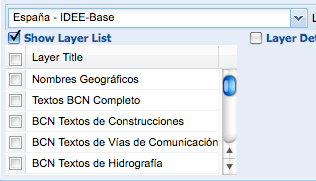
 Some days ago Tim and I had some discussions about how to provide OGC services for biodiversity databases, like for example the Global Registry of Migratory Species. This time the reason for the discussion to start again was the discovery of the new INSPIRE Geoportal Viewer. For those who don't know it, the INSPIRE directive is pushing the creation of a common infrastructure fo sharing geospatial data within Europe. They plan to do that by using Open Standards like the ones from the Open Geospatial Consortium (OGC).
Some days ago Tim and I had some discussions about how to provide OGC services for biodiversity databases, like for example the Global Registry of Migratory Species. This time the reason for the discussion to start again was the discovery of the new INSPIRE Geoportal Viewer. For those who don't know it, the INSPIRE directive is pushing the creation of a common infrastructure fo sharing geospatial data within Europe. They plan to do that by using Open Standards like the ones from the Open Geospatial Consortium (OGC).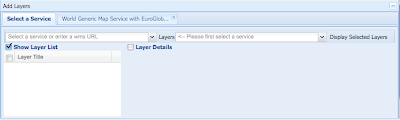
Thursday, June 25, 2009
RSS feeds used by publishers
I am trying to put some recommendations together for publishers on how to encode their RSS feeds or to use other formats to make their digital publications discoverable. If you have any recommendations I'd be glad to know about them. Especially on how to best promote back catalogues of all available publications would be interesting, as RSS feeds natively only show the latest ones (there are paging extensions for Atom, but that has no widespread support). Sitemaps or OAI-PMH seem like a good candidate, although something easier than OAI would be preferred.
Wondering which RSS format is most widely used by publishers currently and which extensions they use to encode their metadata, I wrote a little tool today that reads all current feeds known to ubio and checks their rss format, here are the results, not analyzing the namespaces and extension formats yet:
rss_0.92 = 3
rss_1.0 = 336
rss_2.0 = 431
rss_0.91U = 6
atom_1.0 = 2
So clearly the rdf based rss 1.0 (often together with Prism) and the simple rss 2.0 format is used mostly.
If there only would be a simple way to page. Maybe Microsofts Simple Sharing Extensions could help?
Visualizing Tweeter biodiversity observations
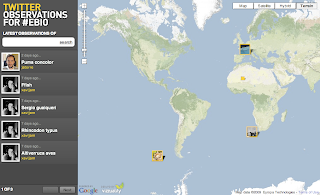 At Ebiosphere09, there was an Informatics Challenge that Rod Page won, congratulations!
At Ebiosphere09, there was an Informatics Challenge that Rod Page won, congratulations!Monday, June 8, 2009
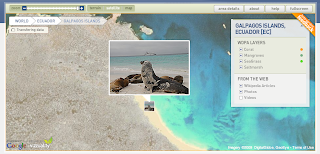
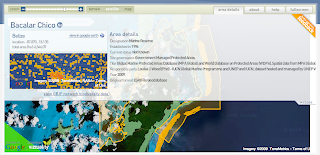
World Database on Marine Protected Areas new website
 The UNEP-World Conservation Monitoring Centre (UNEP-WCMC) today unveiled The World Database on Marine Protected Areas - a site designed to provide the most comprehensive set of Marine Protected Areas (MPAs) available.
The UNEP-World Conservation Monitoring Centre (UNEP-WCMC) today unveiled The World Database on Marine Protected Areas - a site designed to provide the most comprehensive set of Marine Protected Areas (MPAs) available.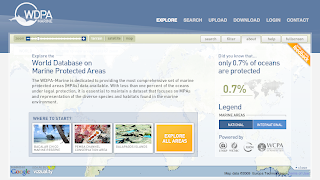
- Tile Mouse Over: To change the cursor when hovering over features on tiles.
- WMS overlays: Dynamically changing the Tile Overlays based on zoom levels for cached and not cached tiles.
- Panoramio and Wikipedia markers without proxies.
- Encoded Polilynes for multipolygons with inner rings.
Saturday, June 6, 2009
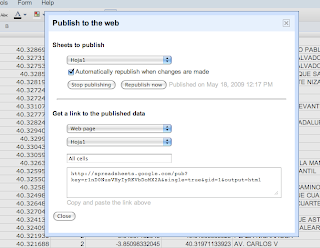
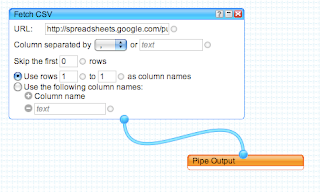
Using Google Spreadsheets with Google Maps for Flash
Thursday, June 4, 2009
Greenpeace BlackPixel | Beautiful Idea
Friday, May 29, 2009
We are going to E-biosphere 09
 E-Biosphere 09 starts next Monday 1st of June on London. Today we want to announce that most of us, Tim Robertson, Sergio Alvarez, Dave Martin and Javier de la Torre, will be there.
E-Biosphere 09 starts next Monday 1st of June on London. Today we want to announce that most of us, Tim Robertson, Sergio Alvarez, Dave Martin and Javier de la Torre, will be there. A new contributor to Biodivertido
My name is Andrew Hill. I am working on my PhD in Rob Guralnick’s lab in the Ecology and Evolutionary Biology department at the University of Colorado. I began working here almost 3 years ago on a project to reconstruct the evolution and spread of influenza through time and over geography (see here or here). Like the influenza itself, my interests have mutated rapidly. Most of my work now involves at least one of three informatics subtypes; biodiversity informatics, phyloinformatics, or (I’m not sure people say this) visual informatics. I have ongoing research on the evolution of influenza, as well as a handful of smaller genomics and bioinformatics projects, and am just now starting work for the OBIS-USA project.
As for my role at Biodivertido, I don’t know yet. Rob, Walter Jetz, and EOL just put on an excellent workshop in Chicago that gave me, Tim, and Javier the opportunity to talk about our interests and coordinate some of our efforts on a couple of new projects. We will likely be giving you more details about some of those in a few hours. So don’t go far from your RSS feed.
For now, all I can say is that I’m extremely excited to be working with these guys and will hopefully be contributing cool things very soon.
Wednesday, April 29, 2009
GBIF-World Database on Protected Areas project
Tuesday, April 14, 2009
WMS Tiling Clients
 For projects relying on open source software and using WMS services for their map applications perhaps the most obvious choice for a client application is to write something using the OpenLayers API. This is a very extensive javascript API and is used to demonstrate the open source OGC compliant servers MapServer and GeoServer. Its straight forward to construct a map and then to overlay multiple layers using WMS services.
For projects relying on open source software and using WMS services for their map applications perhaps the most obvious choice for a client application is to write something using the OpenLayers API. This is a very extensive javascript API and is used to demonstrate the open source OGC compliant servers MapServer and GeoServer. Its straight forward to construct a map and then to overlay multiple layers using WMS services.The screenshot above shows a layer of 0.1 degree cell densities representing data from Australian National Herbarium rendered using GeoServer layered on top of the Google satellite base layer, all pulled together using OpenLayers.
Running this locally the map was rendered by OpenLayers rather quickly. Running this on an externally hosted server I began to notice curious loading of tiles by OpenLayers. So I did a stripped down comparison of the same functionality using the Google maps API. Rendering the cell density tiles from a WMS service in Google maps was done using a javascript function written by John Deck - available here.
The performance improvement with Google maps was immediately obvious. Using the Firefox YSlow plugin we see:
| OpenLayers | Google Maps API |
 |  |
So the performance difference between Google Maps and OpenLayers could be accounted for by any combination of the following:
- Incorrect use of the API (test for OpenLayers is here) and we are missing configuration to reduce tile loading in openlayers
- The tile loading algorithm for Google maps is more efficient in only loading the required tiles for selected view areas
- Openlayers is preloading more images to speed up panning. This would be good, but OpenLayers doesnt seem to prioritise the loading of currently viewed tiles.
Thursday, April 2, 2009
The Taxonomy browser visualization #1 - Tree Lists
 Hi, my name is Sergio Alvarez Leiva and this is my first pos at biodivertido, Finally. I'm part of Vizzuality and work as Interaction Designer for GBIF among others. I will mainly be posting about Interaction Design, UX and Design in general, althought I'll try to post about Front.end developing a little. I hope you find it interesting.
Hi, my name is Sergio Alvarez Leiva and this is my first pos at biodivertido, Finally. I'm part of Vizzuality and work as Interaction Designer for GBIF among others. I will mainly be posting about Interaction Design, UX and Design in general, althought I'll try to post about Front.end developing a little. I hope you find it interesting.Since I began designing for Biodiversity data, I've encountered lot of interesting challenges related to the size of the datasets .
Monday, March 30, 2009
SpatialKey and biodiversity primary data analysis

- The heatmaps are just gorgeous. I would love to know how they do it.
- The timeline filter is great. Has some usability issues but is great.
- The way grids are displayed for summaries. The hover effect is very good and the tooltip very clear.
- The filter "pods" are nice, but I wonder what would happen when you have thousands of hundreds records to search or select on. I suppose that when there is lot of data only the search would be enabled and not the selection.
- Great look and feel.
- Is it necessary to refresh on every map movement? I understand it is on the zoom and if you have the filter by visible area disabled.
- Not having the possibility right now to share the reports as widgets to embed on the blog.
- It would be nice to also let the user provide a polygon or geometry to define the boundaries of the analysis. In this case for example would help a lot to visualize the borders of the protected area.
- The heatmaps!
- The data structures they use for dynamically regrouping the data on the client.
- If it is true that they can handle millions of records, how does the server infrastructure looks like. I know it is Java, but what about the data store, how can they handle the creation of dynamic indexes or how do they do it? Would it work with GBIF data?
Friday, March 20, 2009
How many zoom levels are enough?
We make use of the tiling mechanism employed by many mapping clients, who request 256x256 pixel tiles and then we process the data to be several zoom levels ahead of the one displayed. It is really quite simple, and best described with a couple of examples.
Tuesday, February 24, 2009
Grid data shared as point data. Errors and visualization problems
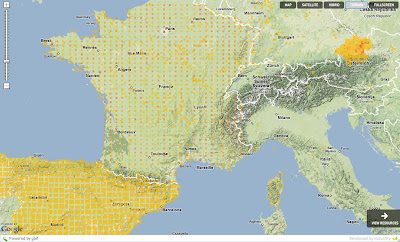
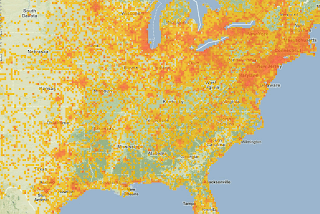
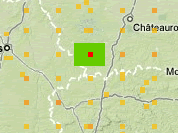
PROJCS["NAD_1983_UTM_Zone_10N",and then they will have to share the easting and northing, like 630084m east, 4833438m north.
GEOGCS["GCS_North_American_1983",
DATUM["D_North_American_1983",SPHEROID["GRS_1980",6378137,298.257222101]],
PRIMEM["Greenwich",0],UNIT["Degree",0.0174532925199433]],
PROJECTION["Transverse_Mercator"],PARAMETER["False_Easting",500000.0],
PARAMETER["False_Northing",0.0],PARAMETER["Central_Meridian",-123.0],
PARAMETER["Scale_Factor",0.9996],PARAMETER["Latitude_of_Origin",0.0],
UNIT["Meter",1.0]]
I was part of the ABCD authors but at this time never looked much into the geospatial part of it. Now it looks to me that this is the case for a correct use of the "variable atomization" method that I did not like later on. Well, then there are cases when I like it. Of couse still work needs to be done on those concepts, and more important, people should start using them!
/DataSets/DataSet/Units/Unit/Gathering/SiteCoordinateSets/SiteCoordinates/CoordinatesUTM
/DataSets/DataSet/Units/Unit/Gathering/SiteCoordinateSets/SiteCoordinates/CoordinatesUTM/UTMZone
/DataSets/DataSet/Units/Unit/Gathering/SiteCoordinateSets/SiteCoordinates/CoordinatesUTM/UTMEasting
/DataSets/DataSet/Units/Unit/Gathering/SiteCoordinateSets/SiteCoordinates/CoordinatesUTM/UTMNorthing
/DataSets/DataSet/Units/Unit/Gathering/SiteCoordinateSets/SiteCoordinates/CoordinatesUTM/UTMText
/DataSets/DataSet/Units/Unit/Gathering/SiteCoordinateSets/SiteCoordinates/CoordinatesGrid
/DataSets/DataSet/Units/Unit/Gathering/SiteCoordinateSets/SiteCoordinates/CoordinatesGrid/GridCellSystem
/DataSets/DataSet/Units/Unit/Gathering/SiteCoordinateSets/SiteCoordinates/CoordinatesGrid/GridCellCode
/DataSets/DataSet/Units/Unit/Gathering/SiteCoordinateSets/SiteCoordinates/CoordinatesGrid/GridQualifier









